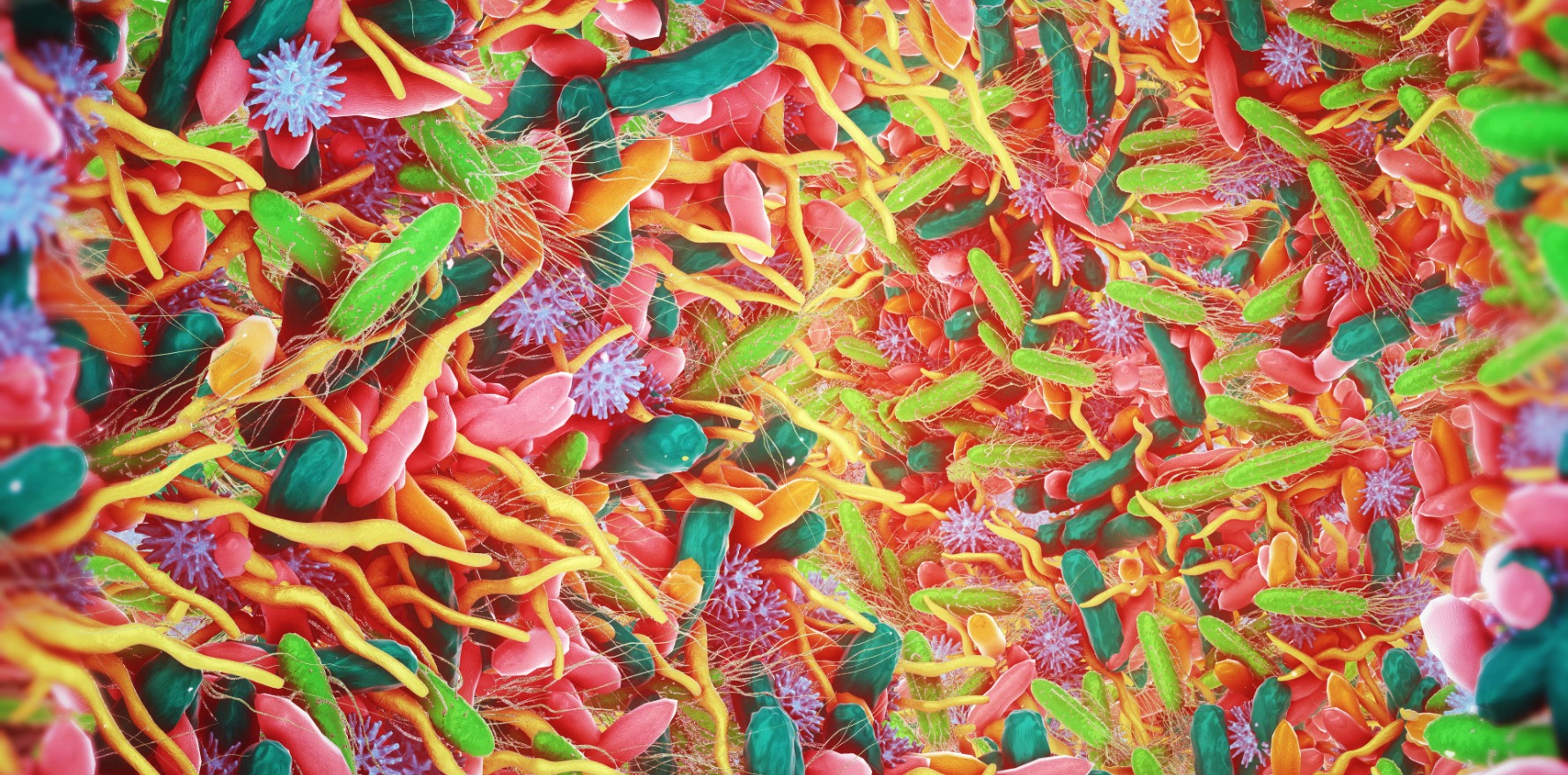
Their discovery has the potential to change how treatments manipulate the microbiome to improve gut and overall health.
An Australian-led international collaboration has proven the feasibility of using culture-based approaches to learn more about phage biology while also identifying a suite of new phage-host pairs.
The human gut contains – among other microorganisms – an incredible number of viruses that infect and replicate within bacteria.
These viruses, known as phages, are believed to influence the makeup of the gut microbiota through multiple different mechanisms, including lysogenic conversion (where they acquire new traits due to its genome being altered after incorporating a phage).
Researchers currently use computational modelling to identify different kinds of phages but have historically lacked a way to determine which and how many of these phages re-enter lytic replication.
Now, a world-first study has used a large-scale, culture-based approach to locate and study temperate bacteriophages in the human gut, resulting in the discovery of more than 100 new viruses. The study findings were published in Nature.
Professor Jeremy Barr, head of the bacteriophage biology research group at Monash University and senior author of the new Nature research, said the findings would significantly change the way researchers think about and study viruses in the human gut.
“We found that compounds produced in human gut cells can wake up dormant viruses inside gut bacteria. This could have major implications for gut diseases like inflammatory bowel disease (IBD), where inflammation and cell death are common,” he told media.
“This work lays the groundwork for future applications in synthetic biology, biotechnology and microbiome therapeutics; it’s a major step forward in decoding the viral dark matter of the human gut.”
Researchers used 252 bacterial isolates from the human gut (93 Bacteroidota, 57 Pseudomonadoata, 51 Bacillota, 50 Actinomycetota and one Fusobacteriota) and a series of computational techniques to identify and experimentally validate inducible prophages.
Each bacterial isolate was exposed to a variety of different experimental induction conditions before being processed for DNA extraction. Although the computations predicted that more than 90% of the isolates would contain a high-quality bacteriophage genome (prophage) region, only 18% of the prophages could be induced.
Related
Further testing using cultured isolates revealed that human host-associated cellular products may act as induction agents, thereby “providing a potential link between gastrointestinal cell lysis and temperate phage populations”.
“Our study highlights the importance of culture-based techniques, alongside experimental validation, genomics and computational prediction, to understand the biology and function of temperate phages in the human gut microbiome. These culture-based approaches will enable applications across synthetic biology, biotechnology and microbiome fields,” the researchers concluded.
Associate Professor Sam Forster, head of the microbiota and systems biology research group at the Hudson Institute of Medical Research and co-author on the paper, was excited about the potential health translation implications of the recent findings.
“Being able to grow these viruses allows us to understand their function and provides the opportunity to develop microbiome therapeutics for diseases from inflammatory bowel disease to cancers,” he said in a statement.
This technology also provides a capacity to engineer probiotic strains with tailored viral functions.”